| TreeGenes: https://treegenesdb.org |
Registrations |
TreeGenes
|
| Tripal |
Basic page |
|
| Tripal v0.3 released! |
Announcement |
Tripal v0.3b has been released and provides the following features:
- Web-based Data Loaders
- Web-based Chado Installation
- Support for the Chado Stock Module
- Support for Feature/Stock Properties, Synonyms, Database References and Relationships
- Integration of Tripal with Drupal Views
- Fully customizable Page Layouts (Little to No HTML or PHP needed)
- Tripal API
More Details
|
| Tripal v3 |
Basic page |

Ontology Driven Design
Controlled vocbularies and ontologies become a central organizing feature of Tripal 3.0 ensuring that biological rather than chado-centric terminology is used. This makes Tripal more comfortable to the biologist for searching and browsing, as well as, more intuitive to the site administrator. Tripal version 3.0 will use Drupal Entities to create ontology-based pages that provide content unique to genes, analyses, maps, libraries, germplasm, publications, etc.
Ease of Customization
Because every biological data type published by Tripal 3.0 is a Drupal Entity, the Drupal web interface allows site administrators to customize data entry forms and display pages. While Drupal core allows you to rearrange fields, customize entry widgets and basic display, contributed modules such as Display Suite and Panels provide complete control over layout allowing you to arrange fields into tables, multiple columns, accordion displays, etc. This greatly reduces the need to customize PHP templates and leads to less coding!
Chado and Beyond
Chado remains the primary database back-end for Tripal and is the ideal location to store most of your biological data. However, the need has arisen for making data stored in other ways (such as BAM, VCF or NoSQL options) also available through your Tripal site. Drupal Entities allow you to add new custom "fields" to your pages and thus aggregate data from Chado with other storage formats. The Drupal and Tripal API provides the programmatic access to create these field and share them with others.
Simplified Administration
Tripal 3.0 will have a simplified administrative interface for all biological content. Your data in Chado is already categorized by ontology terms, therefore, administrators can simply choose to publish the terms defining the data you would like to share. Tripal will publish all data of the specified type, and automatically associate ancillary data linked to it within Chado. Access to private data can be controlled at a granular level such that subsets of data can specified and group permissions set.
Drupal Search API Integration
Since Tripal 3.0 publishes all of your data as entities, it benefits from built-in Drupal Search API integration. This means that all the fields displayed on your Tripal pages will be indexed for searching through a user-friendly, site-wide search box. Furthermore, the Search API supports both Apache Solr and Elastic Search either of which vastly improves performance and provides faceted search allowing users to fine-tune their search results.
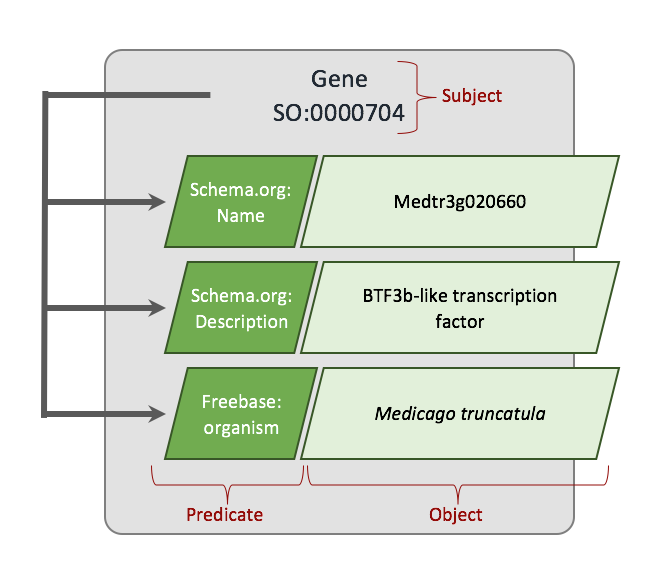
The Semantic Web
All the data presented by Tripal 3.0 will be fully described using the subject > predicate > object tuples required by the Semantic Web. This ensures your data is described in such a way as to be exchanged between communities. Drupal RDFa and XHTML will be used to embed the semantic web tuples in your pages to allow advanced search engines to understand the data presented.
Web Services
All data presented on a page in Tripal 3.0 becomes available via Web services. RESTful Web services will be encoded in JSON-LD which provides an intuitive interface for consumers of web services familiar with JSON. JSON-LD also provides the tuples so important for the semantic web ensuring your content is fully described.
Support for all Data Types
This change in Tripal 3.0 to an ontology-centric design will ensure that if your data is described by an ontology term, even a custom in-house term, you can create pages and attach data. This provides even more flexibility to site administrators!
Million Dollar Question: Backwards Compatibility?
As always, backwards compatibility is a major factor taken into the design of any changes to Tripal. With the change from Nodes to Entities a certain amount of backwards compatibility is lost. Specifically any customizations made to node types, such as custom form elements, will need refactoring. That said, the entire Tripal API will remain backwards compatible and an effort is being made to ensure your custom templates will still work. This, coupled with a mostly automated upgrade path should ease your adoption of Tripal 3.0.
One final consideration is that, although your custom page templates may still work in Tripal 3.0, we encourage you to update your pages to use the new user-interface as this will ensure a more seamless upgrade to Dupal 8. This is due to the fact that Drupal 8 uses the new Twig theme system which requires a complete a rewrite of your current templates.
|
| Tripal 2.0-rc1 Released |
Announcement |
This release candidate provides one last opportunity for bug fixes and minor functional changes prior to the stable release of Tripal 2.0. User's are encouraged to upgrade from Tripal 2.0-alpha to this version.
|
| Tripal @ PAG |
Announcement |
The Tripal community will have several activities at the Plant and Animal Genome (PAG) Conference this weekend! If you will be attending PAG please consider joining us for any of these activities. Below is the list of items:
1) Web Services Meeting: There will be an open meeting to discuss the design and development of Web Services for Tripal/Chado and biological databases in general. The meeting will be held on Saturday at 1pm (my apologies to those who cannot meet this time). Anyone is welcome to attend. Room assignment is yet to be determined.
2) Dinner: An informal dinner is scheduled at the True Food Kitchen restaurant on Saturday night Jan 10th. We will meet in the foyer of the Gold Pacific Ballroom at 6:30 to walk over as a group. The restaurant is located in the bottom floor of the Fashion Valley Mall adjacent to the Town and Country. The restaurant is located next to Bloomingdales.
3) Tripal Workshop: On Sunday at 4pm-6:10pm in the California room we will have the Tripal Workshop open to all PAG attendees. We have several speakers from various groups that are using Tripal. They will provide their experiences as well as introduce work they are doing that could have benefit to all Tripal users. We encourage anyone interested in Tripal, or collaborative development to attend.
|
| Tripal Advisory Commitee Meeting 2020/04/07 |
Tripal Community Meeting Notes |
|
| Tripal Advisory Commitee Meeting 2020/07/07 |
Tripal Community Meeting Notes |
- White paper draft for review: t
- Two community surveys - July 16th deadline
- One anonymous (What do you like? What do you hate? What is the future of tripal?): https://forms.gle/4ri3vWtK1PxNxL9z9
- One non-anonymous (For each DB, data types, funding, extension modules, unique users from the past year): https://forms.gle/RqywukroimvT7Atg9
|
| Tripal Advisory Committee Meeting 2020/10/06 |
Tripal Community Meeting Notes |
Community Highlights
|
Name
|
Highlight Description
|
Can we tweet it? Yes/No
|
|
Lacey-Anne Sanderson
|
ND Genotypes received the Tripal GOLD Badge
|
Yes
|
| |
|
|
| |
|
|
| |
|
|
Agenda
- Hacktoberfest
- Details
- T-shirts and stickers!

- No more January in San Diego. What to do about our annual meetup?
- Proposal:
- Coordinate a virtual Hackathon (Stephen volunteered, anyone else want to help? Jill)
- Flexible timing to work across time zones - a self-organizing hackathon by zoom + slack on times that work best for the groups who want to work on different items.
- Provide some bounded hours - late morning in US for example, to create cohesiveness for break out groups
- Set aside one week and organize group sessions that work for each group during the week.
- Groups could rotate throughout the day
- Topics -
- Tons of T4 options! Fields will be done, will need importers, can pick functionality particularly useful for you, don’t forget extension modules! Working together to upgrade can be a powerful way to develop
- Have a final meeting at a time that works for folks to do reports on those efforts
- Doodle poll for scheduling? Second week of January!
- Here's a site with 8 tips for running a virtual hackathon: https://opensource.com/article/20/8/virtual-hackathon
- Tripal White Paper
- Ready for community review by October 20th:
- Draft: https://docs.google.com/document/d/1w24HzOBJFKG4q3kvgAGwpEWEBgw9773jTcIqMp7bxsg/edit?usp=sharing
- Folder with supplemental files and draft:
https://drive.google.com/drive/folders/1AFSZxWPdcB6fCItdsZCJx87c9s3iErAi?usp=sharing
- Tripal 4 Updates
- Lacey - hoping for alpha by Sept 30 -delay related to fields, fields in 8/9 are quite different. We’d like to make it friendly for extension modules. 8/9 has moved information into “annotations” ie. formatted comments at the top of classes. (related to symphony). The plan is to adopt an additional (optional) software called Drupal Console. Very cool for not only fields but many admin tasks. Drupal console will accept a T3 field and spit out a T4 field (yea!)
- Aiming for alpha in a few weeks. Everyone will need time to upgrade before losing security support in November 2022. Aiming to catch up with existing timeline: https://time.graphics/line/387503
- Plan is for everyone to go from D7 straight to D9 (9.0 and 8.9 testing). Everything in T4 is compatible with D9
- Repo: t4d8 - please go look at the repo and timeline, think about how you are going to tackle this for your database and your extension modules
- GFF Loader - much improved! (perhaps one bug left) - can load an entire genome in just a few minutes
- Functional testing needed - please help!
- Will get rolled into T4
- Deleting features
- Not available in Tripal 3
- recent merge: https://github.com/tripal/tripal/pull/1061
- Good for deleting/unpublishing a small number of entities
- Staton lab: Feature Obliviator
- This Tripal 3 module provides a drush command that allows users to delete data from the chado.feature table by specifying the organism and the analysis for which features should be deleted.
- Command line only - no admin interface
- Depends on Tripal Manage Analyses
- This module creates browseable analysis and library lists for organisms.
- It also provides the obi__organism linker. This linker attaches to analyses and lists organisms linked to it.
- Sean will be expanding to work with other feature types
- JBrowse fields for gene and mRNA pages: (not official, somewhat hacky) https://github.com/NAL-i5K/gene_jbrowse_field
|
| Tripal Advisory Committee Meeting 2021/02/1 |
Tripal Community Meeting Notes |
Agenda
- Update on the Tripal Codefest
- 19 participating, 25 registered (JBrowse will join next time!)
- Breeders API -
- Tripal 4 - 9 PRs!
- Tripal Galaxy
- Gene Pages
- Please fill out survey if you participated in codefest!
- PMC - weekly development meeting will be more in person coding, all welcome to join in
- 3.5 with many bug fixes close to release
- Tripal White Paper
- We received USDA approval last week, its going in this week
- Tripal Grants and Priority Development
- Jill
- CartograTree - (becoming CartograPlant!) - natural populations with georeferenced coordinates, adding environmental layers, Galaxy workflows, visualization, genotypes + phenotypes for landscape genomics
- TreeSnap mobile app
- Genome Annotation - deep learning approaches - functional info + RNA folding - at the end of the grant (3ish years) move the workflow in Tripal
- Methylation proposal - perennial species - best practices for comparing across data sets, how to integrate into Tripal
- Gene families and orthogroups
- Pangenomes
- Core Tripal!
- Focus on solid use cases
- Stephen
- NSF - SciDAS - workflows on the cloud - RNASeq particularly
- Integrate tripal workflows (nextflow, easy to dockerize, run on kubernetes or slurm or PBS or other)
- Future nextflow tripal module?
- Working on tripal file module - ready for testing!
- license/usage
- associate files to data types
- Make files available through web services
- NRSP10 (PI Dorrie Main)
- Tripal core development and outreach
- Full time - Josh coming to Tripal this month
- Part time - Risharde
- Multiomics data integration, metagenomics, microbiomes, all the omes!
- Network module - close!
- Docker - a popular solution for dev and production Tripal sites
- Singularity - supports docker images and friendlier on HPCs
- Lacey - her group uses Docker entirely for development and for production in one project
- Monica - moving toward docker for production
- This might make a great future dev or user meeting topic!
- Main Lab (Sook)
- NRSP10 - core (see above) Oct 2019 - Sept 2024
- 3 developers for
- epigenome module
- Map viewer w/ genome view
- TripalBIMS themes to support phenomics data and BrAPI compatibility and GWAS tools
- Curation module
- Preproposal - USDA SCRI - application in
- Curation of data
- 1 developer for modules - pangenome module, map viewer, SNP genotype and haplotype module, synteny viewer and Community gene curation - enable community members to contribute (like TAIR GOAT)
- Focus on linking module to facilitate more integrated display
- Funding for Training and outreach
- PGRP - pending
- Project database for rice
- In discussion for funding support for developing a bacterial database for Metabolomic and proteomic data
- USDA - consistent messaging from plant and animal communities about database support -
- Team up with Fiona (idea from Monica)
- Lacey
- Genome Canada
- Evolves - not just Tripal, KnowPulse and resources
- Integration of 3rd party tools - JBrowse2, SynVisio, Genotype matrix tool to support haplotypes and haplotype views
- EVOLVES Project page: knowpulse.usask.ca/evolves
- We are beginning year 3 of a 4 year grant
- DivSeek Canada pilot project - focused more generally (not just pulses)
- https://www.divseekcanada.ca/
- Completely focused on Tripal
- Docker container for breeding/variant focused Tripal site, already configured, Tripal Crop Docker, first version done
- https://www.divseekcanada.ca/tripal-crop-docker/
- Funding covers documentation, already has an administrative guide, videos showing functionality
- Image -> container -> script pulls down all modules based on an environmental file -> run configure w/ crop info -> modules are configured automagically
- Pilot portal for smaller crops can use this as a central repo without having to maintain their own site
- Example data already loaded for 3 species
- https://staging.divseekcanada.ca/
- Lots of outreach to breeding/variant focused community in Canada
- Our group is focused on third party tool integration, structural variation, pangenome, and breeder support, as well as, outreach
- Monica
- Sean Buehler - funded by USDA cooperative agreement
- AgBioData - RCN -
- Identify priority issues
- Include stakeholders and funders
- Education component - FAIR data
- Sustainability analysis - how databases can continue to exist with more data but less funding (Phoenix Bioinformatics)
- Comparative genomics, gene family display, building on legume community (Valentin - greenphyl.org - not Tripal, maybe one day :)
- Valentin
- Moving from 2 to 4 (skip 3!)
- Moving drupal 7 to 9
- Less reliance on chado - nice that its normalized and standard but complicated queries are a problem and not fast - materialized views or drupal solutions (data in Drupal sync with Chado data) may be implemented (or other approaches… still in progress)
- Chado for loading but not querying
- Banana genome hub v2 on T3 just released! https://banana-genome-hub.southgreen.fr/
- New modules - plant genome viewer, next year
- BrAPI module - need to move to T4 - no funding for Valentin but can work with Lacey
- Rodrigo - AgriFood Canada - PI Genome Alberta project
- Bee microbiome
- Switzerland, Germany, large international group!
- Just began, beta version of portal developed - considering adopting Tripal
-
- Opportunity to collaborate or build on resources from AGR
|